E. coli Model for Protein Amplification
Escherichia coli is a bacterium that most of us know first from recalls of foods like meat and lettuce. Although E. coli is a commensal (non-harmful) gut bacterium, the wrong circumstance and strain can make infection uncomfortable and dangerous. E. coli is gram negative, meaning it has a double-layered cell membrane that makes it more difficult to treat with antibiotics.
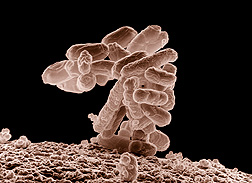
Our homogenization equipment is regularly used to disrupt E. coli. In “Identification and Biosynthesis of pro-inflammatory sulfonolipids” authors Hou et al. use the NanoGenizer to break apart E. coli after using it for protein expression. Many other researchers have done the same (https://www.genizer.com/u_file/2206/file/6bffa6a456.pdf, https://www.genizer.com/u_file/2312/file/298f8a65e6.pdf ,
https://www.genizer.com/u_file/2312/file/3ca3c35d8a.pdf)
What makes this common cause of diarrheal illnesses a welcome tool in so many research labs?
First, we need to understand the research problem E. coli is being used to address in many labs.
Researchers often need large amounts of proteins for a variety of purposes. One common example is for pharmaceutical production at a large scale. However, having to obtain that protein from its natural source can be time-consuming, resource intensive and complicated. Instead, researchers will often take the gene that encodes that protein and transform it into a bacterium. There, the bacteria will make that protein more quickly. However, not just any bacteria are suited for this purpose, and that’s what makes E. coli a laboratory star.
So researchers, go forth and enjoy this model organism in the laboratory, if not always in your kitchen!
Sources:
https://www.ncbi.nlm.nih.gov/books/NBK564298/
https://pmc.ncbi.nlm.nih.gov/articles/PMC4029002/
https://www.ncbi.nlm.nih.gov/books/NBK562895/

Our homogenization equipment is regularly used to disrupt E. coli. In “Identification and Biosynthesis of pro-inflammatory sulfonolipids” authors Hou et al. use the NanoGenizer to break apart E. coli after using it for protein expression. Many other researchers have done the same (https://www.genizer.com/u_file/2206/file/6bffa6a456.pdf, https://www.genizer.com/u_file/2312/file/298f8a65e6.pdf ,
https://www.genizer.com/u_file/2312/file/3ca3c35d8a.pdf)
What makes this common cause of diarrheal illnesses a welcome tool in so many research labs?
First, we need to understand the research problem E. coli is being used to address in many labs.
Researchers often need large amounts of proteins for a variety of purposes. One common example is for pharmaceutical production at a large scale. However, having to obtain that protein from its natural source can be time-consuming, resource intensive and complicated. Instead, researchers will often take the gene that encodes that protein and transform it into a bacterium. There, the bacteria will make that protein more quickly. However, not just any bacteria are suited for this purpose, and that’s what makes E. coli a laboratory star.
- Speed: E. coli can double as fast as every 20 minutes. Optimal conditions for quickly producing large quantities of protein.
- Density: E. coli can grow densely, reducing the amount of space required for protein production.
- Materials: E. coli can grow well on affordable and easily obtainable materials, making it accessible to researchers.
- Universality: The wide breadth of research done using E. coli as a model makes it easier to work with.
- Tractability: Tractability refers to the ease of editing a genome. E. coli is naturally genetically tractable and can pick up new genes and add them to itself. This eases the process of adjusting its genome for researchers. As a pathogen, this feature also helps E. coli evade antibiotics.
So researchers, go forth and enjoy this model organism in the laboratory, if not always in your kitchen!
Sources:
https://www.ncbi.nlm.nih.gov/books/NBK564298/
https://pmc.ncbi.nlm.nih.gov/articles/PMC4029002/
https://www.ncbi.nlm.nih.gov/books/NBK562895/

 USD
USD 949-932-0294 Los Angeles
949-932-0294 Los Angeles  Nano@Genizer.com
Nano@Genizer.com